Introduction
First-trimester chorionic villus sampling (CVS) is used as an alternative to prenatal invasive diagnosis by amniocentesis. quantitative fluorescent polymerase chain reaction (QF-PCR) for the detection of common autosomal aneuploidies was developed in 1993 [
1] and introduced as a National Health Service diagnostic test in 2000 [
2,
3]. Today, it is a well-established rapid aneuploidy test for prenatal samples obtained by CVS and amniocentesis, and is used in many laboratories as an adjunct to long-term culture (LTC). In most laboratories QF-PCR is widely used on prenatal samples for rapid aneuploidy testing of trisomies 13, 18, and 21, but is usually combined with LTC and conventional karyotyping [
4]. In contrast to QF-PCR, conventional karyotyping is usually performed using in vitro cultures of fetal cells; dividing cells are harvested and metaphase stained by Giemsa, followed by microscopic analysis. Most pregnant women with an increased risk of aneuploidies such as increased fetal nuchal translucency (IFNT) at ultrasound scanning or a positive serum screening test and their family members want rapid aneuploidy testing to lessen their anxiety. Although CVS provides a shift toward first-trimester cytogenetic diagnosis, which is generally performed at 11 to 13 weeks of gestation, the
in vitro cell culture before G-banding staining usually takes 2 weeks. Therefore, pregnant women with an increased risk of chromosomal abnormalities must keep waiting until the final results are confirmed followed by the prenatal invasive procedure, which does not resolve their anxiety. However, of the chromosomal aneuploidies, 78% are numerical variations of chromosomes 13, 18, and 21 [
5], which can be detected by QF-PCR. In addition, QF-PCR provides rapid results following CVS by bypassing culturing of the fetal cells. QF-PCR has been widely used in most developed countries, but the efficacy of CVS with QF-PCR has not been validated in Korea. To date, only a few case reports of complete discrepancies between diagnostic QF-PCR and CVS karyotyping results for common selected autosomal trisomies of chromosomes 13, 18, and 21 have been described [
6,
7,
8,
9] and there have been no reports on QF-PCR by CVS in Korea. Therefore, the primary outcome of this study was the detection rate of the common aneuploidies with trisomies 13, 18, and 21 by QF-PCR via CVS. The secondary outcome was the comparison of results between QF-PCR and conventional cytogenetic results. Additionally, the results of QF-PCR were analyzed according to the indication of CVS.
Materials and methods
We retrospectively reviewed the medical records of consecutive pregnant women who underwent QF-PCR and cytogenetic testing following CVS at Cheil General Hospital between December 2009 and June 2014 for prenatal aneuploidy screening. Only cases of reported QF-PCR before LTC were included in the present study. Before the CVS procedure, a 10-mL volume of maternal blood was collected in an ethilen dianmin acetic acidtube to rule out maternal cell contamination. In our center, CVS was performed with the transabdominal or transcervical approach according to the position of the placenta. After the CVS procedure, two separate villus fronds were set aside independently for rapid QF-PCR aneuploidy testing in order to minimize the risk of misdiagnosis due to confined placental mosaicism. Chorionic villus tissue of approximately 1 to 2 mm in size was cleaned to remove maternal decidua or blood clots and finely chopped. Dissociated villus samples were transferred to a 1.5-mL tube and centrifuged at 13,000 rpm for 1 minute. DNA extraction was achieved by addition of Chelex, InstaGene Matrix (Bio-Rad, Hercules, CA, USA) solution to the pellet after discarding the supernatant. The cell pellet was vortexed and incubated for 10 minutes at 100Ōäā. The samples were centrifuged again at 13,000 rpm for 1 minute, and 2 ┬ĄL of the resultant supernatant was used in the QF-PCR assay. These villus samples were successfully amplified using Aneufast (Genomed AG, Wollerau, Switzerland) kits containing five multiplex markers on each of the chromosomes. This kit uses a five-dye fluorescent system for automated DNA fragment analysis and allows multiplex amplification and electrophoresis of more than 15 loci simultaneously. Two multiplex QF-PCR sets (S1 and S2) are used to perform initial aneuploidy screening and the assays are designed for analysis by capillary electrophoresis. In addition, there are four chromosome-specific extra marker sets (M21, M13, M18, and MXY) that may be used as back-ups in cases where less than one marker (S1/S2) on any one of these chromosomes is informative.
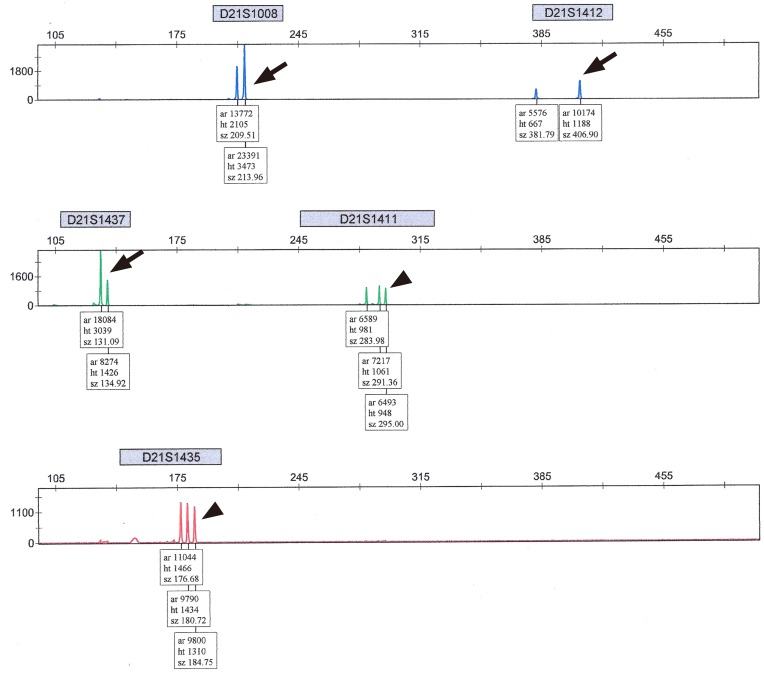
The amplified DNA samples were separated by electrophoresis using an ABI 3130xl Genetic Analyzer, and DNA representing each allele for a specific marker was quantified by its peak using GeneMapper Software ver. 4.0 (Applied Biosystems, Foster City, CA, USA). The interpretation was performed as follows: peak height ratios of allele dosage between 0.8 and 1.4 on at least two informative markers, were defined as normal and reported as negative. Markers with allele ratios between 1.4 and 1.8 or single peaks were interpreted as uninformative (
Fig. 1). The presence of three alleles with an equal peak height ratio (
Fig. 2) or with a ratio of Ōēż0.6 or Ōēź1.8 was considered a trisomy and was reported as positive (
Fig. 3). In our center, we considered positive and uninformative QF-PCR findings as abnormal results, and recommended further confirmation by LTC in such cases. We report the common autosomal trisomies (chromosomes 13, 18, and 21) but have not officially reported abnormalities regarding sex chromosomes determined by QF-PCR assay, waiting for LTC.
In our center, uninformative QF-PCR results were reported in cases of abnormal findings in only a single marker, and these cases were recommended for confirmation by LTC. Additionally, for conventional cytogenetic analysis, monolayer cultures were harvested at 10 and 11 days for each sample. The patient's age at CVS for QF-PCR, parity, body mass index, indications of CVS procedure, gestational age at CVS, reports of QF-PCR assay, reports of cytogenetic study, days from the procedure to QF-PCR results, and days from the procedure to the final report of the cytogenetic study were collected by chart review. Ethical approval of this study was obtained from the institutional review board (CGH-IRB-2015-50) and the need to obtain informed consent was waived. Statistical analysis was conducted using the SPSS ver. 12.0 (SPSS Inc., Chicago, IL, USA). The data are presented as means, standard deviations, or medians (interquartile ranges) for continuous variables and as numbers (percentages) for nominal or categorical variables.
Results
During the study period, QF-PCR assays were performed on 403 (93.5%) of the 431 prenatal chorionic villus samples; the remaining 28 (6.5%) samples were not analyzed concomitantly by QF-PCR. All 403 chorionic villus samples from 383 pregnant women were successfully amplified by PCR.
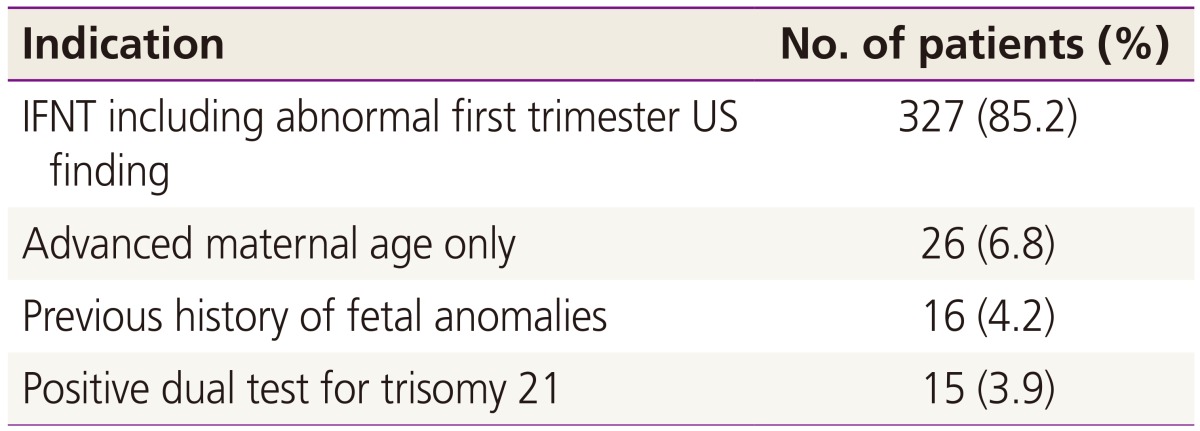
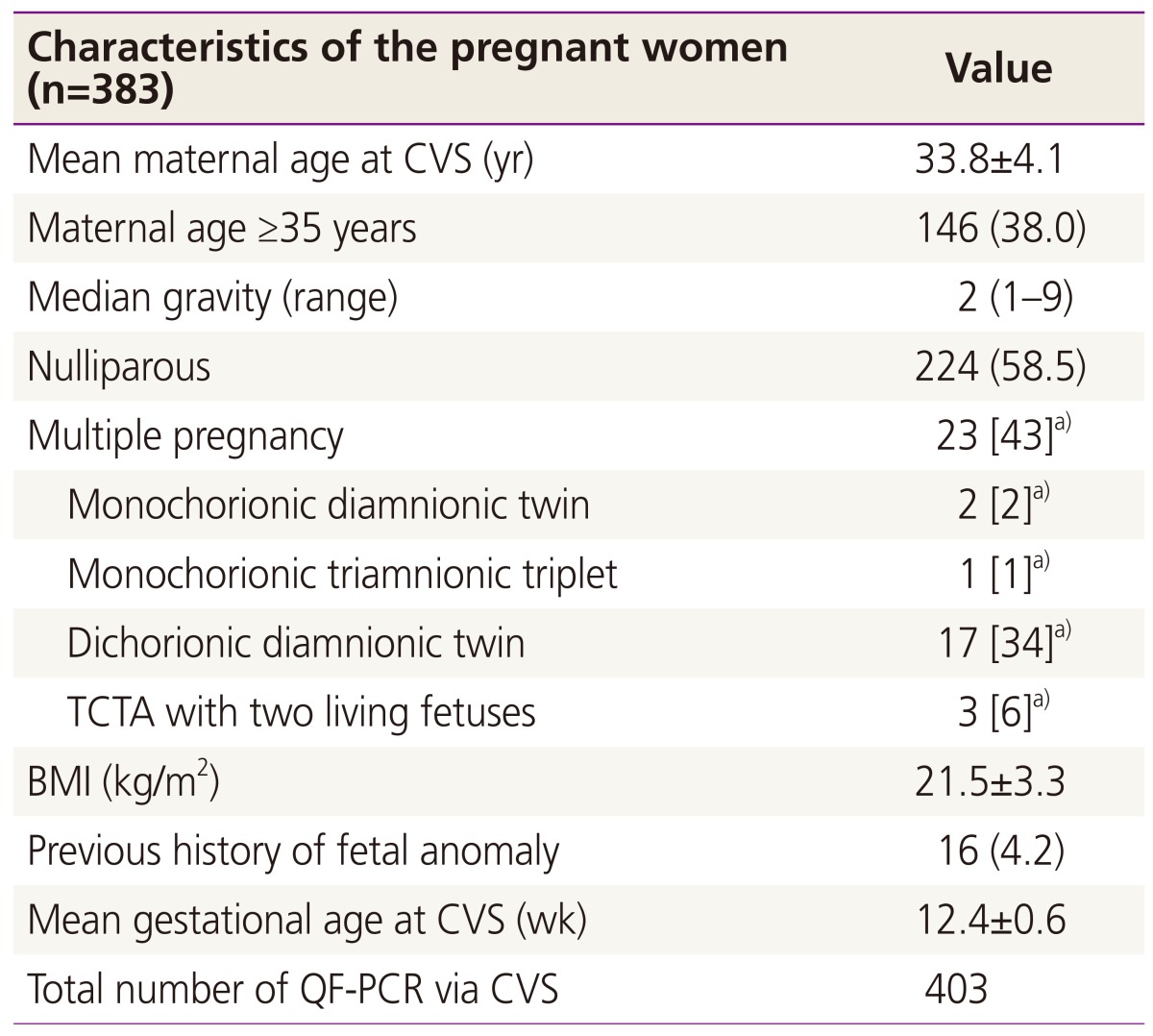
Table 1 shows the maternal characteristics of patients who underwent CVS with QF-PCR. The mean maternal age was 33.8┬▒4.1 years, with 38.1% of the group being older than 35. The median gravity was 2. Among the study population, 227 (59.3%) cases were nulliparous and 23 were multiple gestations. The average gestational age at CVS was 12.4┬▒0.6 weeks. Of these procedures, the transabdominal approach was used in 326 (80.9%) cases and the transcervical approach was used in 77 cases (19.1%). Location of placenta was as follows: 261 anterior, 121 posterior, 16 lateral, and 5 fundus of the uterine cavity. The median number of needle insertions was 1 (range, 1 to 4). It took a median of 1 day for the QF-PCR results to be made available to the mother and genetic counselor, and the final cytogenetic results were reported a median of 14 days after the procedure. Indications of CVS are listed in
Table 2 [
10]. The major indications for QF-PCR were an abnormal first-trimester ultrasonographic (US) finding including IFNT (326, 85.1%), followed by advanced maternal age alone (26, 6.8%), previous history of fetal anomalies (16, 4.2%), and a positive dual test for trisomy 21 (15, 3.9%). The results of rapid aneuploidy screenings and conventional cytogenetic analyses are reported in
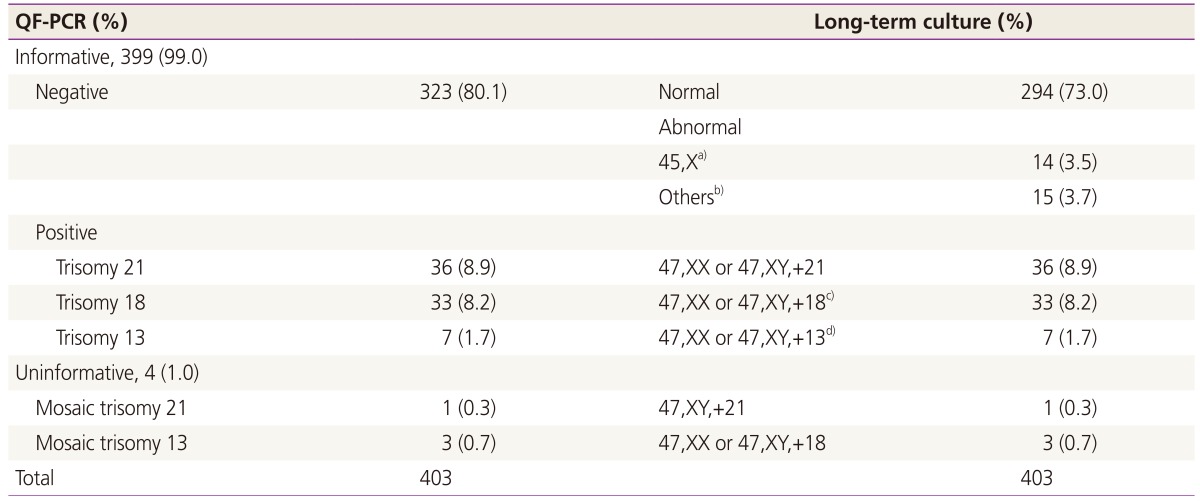
Table 3. Of the 403 samples, 76 cases of chromosomal abnormalities (36 cases of trisomy 21, 33 cases of trisomy 18, and 7 cases of trisomy 13) were detected by QF-PCR. Additionally, four cases (1.0%) of suspected trisomy (one case of mosaic trisomy 21 and three cases of trisomy 18) that were reported as uninformative in QF-PCR were confirmed as the corresponding trisomies by cytogenetic analysis. Thus, 109 cases of chromosomal abnormalities (80 cases of common trisomies, 14 cases of monosomy X, and 15 cases of other structural aberrations of chromosomes) were detected by LTC. Among the common chromosomal trisomies, two cases had microstructural abnormalities; one case of trisomy 18 had a translocation (47,XY, t[3;22][q26.3;q12],+18) and one case of trisomy 13 had derivatives in addition to the autosomal trisomies (46,XX, +13,der[13;13][q10;q10]). The sensitivity of QF-PCR was 100% (80/80) and the specificity was 100% (323/323) for common trisomies 13, 18, and 21, as uninformative findings were regarded as abnormal and recommended for confirmation by LTC in our center.
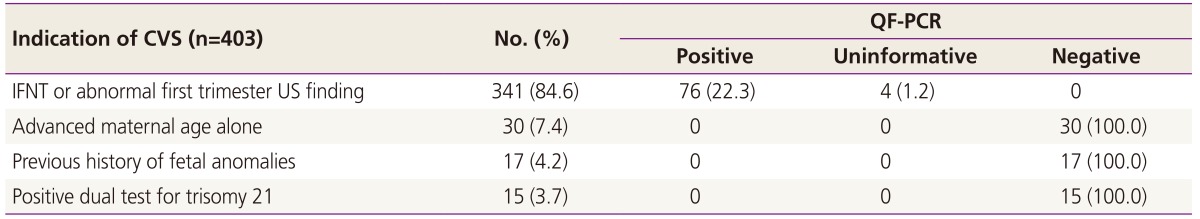
Table 4 shows results of QF-PCR according to indications of CVS. IFNT with or without abnormal US findings was the most common indication (84.6% of QF-PCR cases), and all cases with abnormal QF-PCR results had an identical indication of IFNT prior to CVS.
Discussion
Pregnant women at a high risk of chromosomal abnormalities have the options of CVS, amniocentesis, or cordocentesis as prenatal invasive procedures to extract fetal genomic DNA. Recently, CVS with QF-PCR has been widely used for the rapid diagnosis of aneuploidies in the first trimester [
11]. One of the advantages of CVS over amniocentesis and cordocentesis is that the results are available earlier in the pregnancy [
4]. Although CVS can allow for a first-trimester prenatal invasive diagnosis, most conventional karyotyping takes 2 weeks before reporting as it requires cell cultures [
12]. The combination of QF-PCR with CVS can be helpful for more rapid detection of common autosomal trisomies and reduces parental anxiety earlier when the results are normal. QF-PCR findings can be reported within 24 to 48 hours as
in vitro cell cultures are not required [
6,
13,
14]. In addition, other advantages of QF-PCR are that it only requires a small sample and it also allows automation of the procedures, as it does not require a highly qualified technician [
4]. As a result of these advantages, QF-PCR is now a well-established technique for the rapid testing of samples from prenatal invasive procedures for trisomies 13, 18, and 21, and, in some cases, for sex chromosome abnormalities [
15].
However, QF-PCR on CVS has several limitations. First, mosaicisms can be present in approximately 1% to 2% of the chorionic villus samples, and most of them are confined to the placenta [
16]. Chorionic villus tissue is composed of two cell lineages, chorionic ectoderm (cytotrophoblast) and chorionic mesoderm, both of which are embryologically distinct from the fetus. Even if the placental tissue shows the mosaic pattern, the fetus can be non-mosaic for either diploidy or aneuploidy. In our data, four cases that showed mosaic trisomies 18 and 21 in QF-PCR were confirmed as non-mosaic trisomies 18 and 21 in LTC. Mosaicism of QF-PCR in uncultured chorionic villus samples may be due to confined placenta mosaicism in 1% to 2% of CVS cases [
11,
12,
17], of which only 5% to 25% have true fetal mosaicism [
16,
18]. QF-PCR assays cannot detect mosaicisms that represent less than 15% of the cell population [
19]. Our four cases were reported as uninformative due to suspected mosaicisms. However, we informed the physician of the abnormalities in these cases and recommended further analysis by LTC, which confirmed all four cases as non-mosaic trisomies. This discrepancy may have been due to differences in the methods, as karyotype analysis only examines cells that are in metaphase whereas QF-PCR analysis examines all cells, including non-dividing cells. QF-PCR can detect mosaicisms with low-level trisomy cell lines [
19]. To date, only a few case reports of complete discrepancies between diagnostic QF-PCR and CVS karyotyping results for common selected autosomal trisomies of chromosomes 13, 18, and 21 have been reported [
6,
7,
8,
9,
20]. In the past, the incidence of true false-negative CVS results by short-term culture and LTC was estimated to be lower than 0.03% (1 in 3,300) [
21]. The first report of complete discrepancies between CVS QF-PCR and cytogenetic analysis based on LTC and positive karyotyping in normal QF-PCR was in 2006, and the predominance of normal diploid cytotrophoblasts in the retrieved villi might explain the discrepancies between normal findings in QF-PCR and trisomy 18 in LTC [
6]. We found 2 cases of sex chromosome mosaicism among CVS cases during the study period. In the first case, a 36-year-old pregnant primigravida woman underwent QF-PCR via CVS for the indication of US cystic hygroma suspicious for mosaicism of monosomy X. We consider this case to be true fetal mosaicism because of US findings, and recommended amniocentesis for confirmation. However, she refused and was lost to follow-up. In the second case, a 32-year-old, pregnant gravida 3, para 1, pregnant woman underwent amniocentesis, and 46,XY without mosaicism was confirmed. She delivered a healthy baby.
Second, maternal cell contamination is common in CVS [
17]. Before the QF-PCR assay, we carefully cleaned the retrieved chorionic villi to remove maternal blood clots. We sampled all cases of maternal blood before CVS to prevent maternal cell mosaicism in our center and tested maternal blood in all QF-PCR analyses in CVS to exclude maternal cell mosaicism.
Third, mixed findings can result from polymorphisms or partial chromosome imbalance. Submicroscopic duplications of satellites showing triallelic patterns for a single marker when all markers on a chromosome are normal are rare but can be detected by QF-PCR [
22]. Submicroscopic duplications can be inherited from one normal parent, and are insignificant findings. If submicroscopic duplication was suspected in QF-PCR of CVS samples, we recommended that both parents undergo QF-PCR with blood sampling. We considered that there was no evidence of trisomies of chromosome 13, 18, or 21 in QF-PCR if one of the parents was normal and an identical pattern was shown in the fetus.
Fourth, QF-PCR sensitivity and specificity are dependent on the methodologies used in the laboratories and whether they report abnormalities of sex chromosomes [
9,
12,
20]. The methodology of QF-PCR differs according to the guidelines of the laboratory and the automatic machines used for amplification and analysis. Work in our institution was targeted and reported only selected autosomal trisomies of chromosomes 13, 18, and 21; we have not officially reported on the sex chromosomes, as most abnormalities of sex chromosomes require confirmation by LTC and it can cause unnecessary parental anxiety during the wait for LTC results. This is because QF-PCR has the limitation that it may not detect rearrangements and mosaicism [
23]. Although we have detected many abnormalities in sex chromosome at our center, it is difficult to detect whether the sex chromosomes are mosaicisms or structural abnormalities.
Each case of trisomy 18 and 13 had combined structural abnormalities, such as derivatives or translocations. All women with abnormal QF-PCR results had concordant common autosomal trisomies in LTC. Uninformative findings that had only one abnormal single marker were found in four cases and were suspected mosaicisms consistent with common trisomies. The physicians were informed of suspected mosaicisms consisting of each trisomy on QF-PCR, and were recommended to wait for the final results from LTC. All other women were confirmed as having normal fetal karyotypes by LTC except for 29 cases with other non-targeted chromosomal aberrations that could not be detected by QF-PCR or were not reported in our center. These were 15 cases (3.7%) of other structural chromosomal abnormalities and 14 cases (3.5%) of non-mosaic monosomy X. However, most aneuploidies were trisomy 13, 18, or 21. In our data, the sensitivity and specificity of QF-PCR for the common autosomal trisomies were both 100% as uninformative findings were regarded as abnormal and recommended for confirmation by LTC in our center. Up to August 2015, we have performed QF-PCR in a total of 7,943 amniotic fluid samples since early 2007 and 732 CVS samples since November 2008 for the rapid diagnosis of common trisomies of chromosomes 13, 18, and 21. In the present study, we report the results of screening for 403 consecutive chorionic villus samples in our institution using both QF-PCR and conventional cytogenetic analysis between 2009 and 2014.
Our data represent the first reports of QF-PCR in CVS in the Korean population with high sensitivity and specificities for a relatively large number of consecutive pregnant women. Furthermore, there were no complete discrepancies between QF-PCR and LTC. The overall positive rate of aneuploidies involving the selected chromosomes 13, 18, and 21 and excluding the sex chromosomes was 18.9% (76/403), which is higher than that reported in previous studies [
24].
The most common indication (84.9%) for invasive CVS was IFNT with or without abnormal US findings in the first trimester, and 23.4% of abnormal (positive or uninformative) QF-PCR findings via CVS were in this category for the indication of CVS. Our data have a higher positive rate of QF-PCR because we included exclusively pregnant women at high risk for aneuploidies. Although noninvasive prenatal diagnosis with maternal serum cell-free fetal DNA is now possible, the results usually take 2 weeks and positive noninvasive prenatal testing must be confirmed by invasive prenatal diagnosis [
25]. Our study suggests that CVS with QF-PCR is better for pregnant women with IFNT and/or abnormal US findings in the first trimester than other screening tests for fetal aneuploidy.
In conclusion, CVS using QF-PCR is valuable for rapid prenatal screening in pregnant women with a high risk of aneuploidies, especially women with IFNT with or without abnormal US findings in the first trimester. The availability of rapid prenatal aneuploidy detection for common autosomal chromosomal trisomies for all women undergoing an invasive prenatal procedure can alleviate parental anxiety and also reassures parents sooner if the results are not positive for trisomies. To date, our institution has only reported on selective chromosomes 13, 18, and 21 and not on sex chromosomes, and revalidation of short tandem repeats including sex chromosomes will be required through a larger study in the future.