|
 |
- Search
| Obstet Gynecol Sci > Volume 64(5); 2021 > Article |
|
Abstract
Supplementary material
Notes
Ethical approval
This study was approved by the International Medical University (IMU) Joint-Committee on Research and Ethics (BMS I-2019[09]). In accordance with the journal’s guidelines, we will provide our data for the reproducibility of this study in other centers if requested. The study was performed in accordance with the principles of the Declaration of Helsinki.
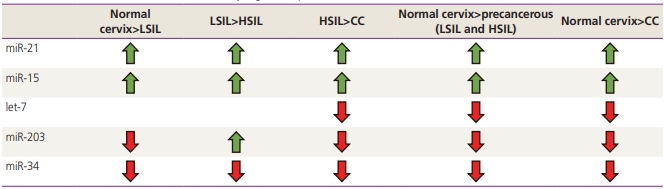
Fig. 1
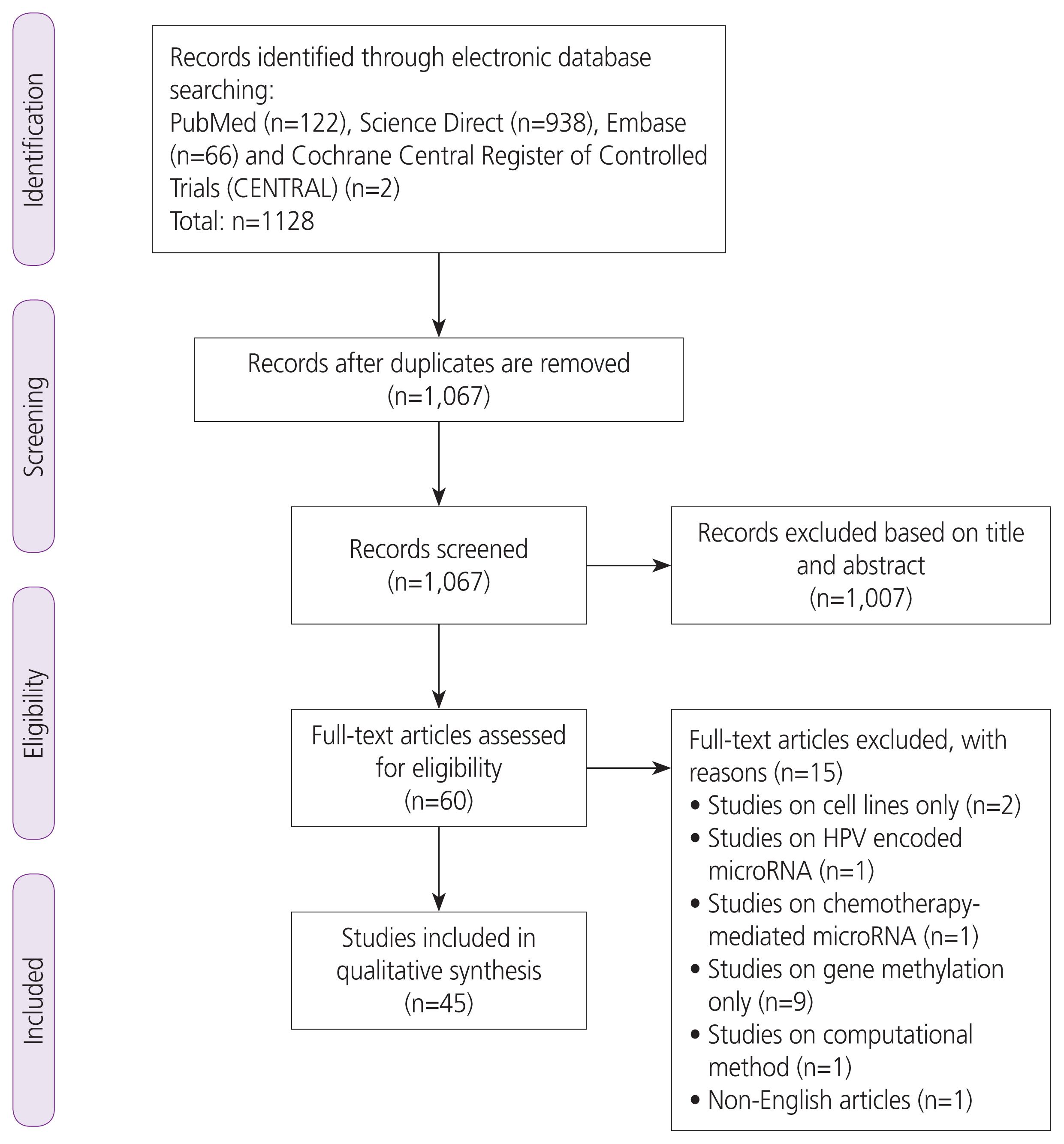
Table 1
Table 2
| Title | Author, year, country, type of study | Sample size, sample type | Age group | Method, reference gene | miRNA mentioned (significant) | Normal cervix to CC | Normal cervix to CIN I-III | Normal cervix to LSIL (CIN I) | LSIL (CIN I) to HSIL (CIN II-III) | HSIL (CIN II-III) to CC | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|
|
|||||||||||
| Upregulated | Downregulated | Upregulated | Downregulated | Upregulated | Downregulated | Upregulated | Downregulated | Upregulated | Downregulated | ||||||
| 1. Oncogenic microRNA signature for early diagnosis of cervical intraepithelial neoplasia and cancer | Liu et al. [39], 2018, Hong Kong, case-control study | 145 NILM, 239 LSIL, 285 HSIL, 58 CC, frozen and FFPE tissue | Mean: NILM (50), LSIL (39), HSIL (42), CC (51) | Microarray analysis and validated by RT-qPCR, RNU6B | Mir-20a, Mir-92a, Mir-141, Mir-183, Mir-210, Mir-944 | Mir-20a, Mir-92a, Mir-141, Mir-183, Mir-210, Mir-944 | Mir-20a, Mir-92a, Mir-141, Mir-183, Mir-210, Mir-944 | Mir-20a, Mir-92a, Mir-141, Mir-183, Mir-210, Mir-944 | Mir-92a, Mir-183 | Mir-20a, Mir-141, Mir-210, Mir-944 | |||||
|
|
|||||||||||||||
| 2. Microrna-551b expression profile in low and high-grade cervical intraepithelial neoplasia | Lukic et al. [30], 2018, Italy, case-control study | 10 Normal, 18 condyloma, 8 CIN I, 14 CIN II-III, FFPE tissue | Mean: 35.98±9 | RT-qPCR, U6snRNA | Mir-551b | Mir-551b | Mir-551b | Mir-551b | |||||||
|
|
|||||||||||||||
| 3. Mir-155-5p inhibits PDK1 and promotes autophagy via the mtor pathway in cervical cancer | Wang et al. [43], 2018, China, case-control study | 48 Normal, 18 CIN I-II, 18 CIN III-CC, fresh tissue | N/A | RT-qPCR, U6snRNA | Mir-155-5p | ||||||||||
|
|
|||||||||||||||
| 4. Comparable expression of Mir-Let-7b, Mir-21, Mir-182, Mir-145, and P53 in serum and cervical cells: diagnostic implications for early detection of cervical lesions | Okoye et al. [35], 2019, Nigeria, cross-sectional study | 159 NILM, 46 cervicitis, 46 ASCUS, 40 LSIL, 28 HSIL, 10 SCC, serum and exfoliated cells | Mean: 39.45±11.16 | PCR and gel electrophoresis, Mir-16 | Mir-21, Mir-146a, Mir-155, Mir-200c, Mir-182, Let-7b, Mir-145 | Mir-21, Mir-146a, Mir-155, Mir-200c | Mir-21, Mir-146a, Mir-182 | Mir-145 | Mir-182, Mir-21, Mir-182, Mir-200c (cell) | Mir-146a, Mir-200c (serum) | Mir-21, Mir-146a, Mir-155, Mir-200, Let-7b | Mir-182 | |||
|
|
|||||||||||||||
| 5. MicroRNA expressions in HPV-Induced cervical dysplasia and cancer | Gocze et al. [31], 2015, Hungary, cross-sectional study | 30 CINI, 10 CIN, 20 CIN III, 38 SCC, FFPE tissue | Mean: 36.17 | RT-qPCR, 5srRNA, U6snRNA | Mir-27a, Mir-34a, Mir-155, Mir-196a, Mir-203 | Mir-27a, Mir-155 | Mir-34a | Mir-27a | Mir-196a, Mir-34a | ||||||
|
|
|||||||||||||||
| 6. Plasma expression of mirna-21, -214, -34a, and -200a in patients with persistent HPV infection and cervical lesions | Yang et al. [45], 2019, China, case-control study | 73 Normal, 19 CIN I, 54 CIN II, 71 CIN III, 15 CC, serum and FFPE tissue | Mean: normal (40.94±7.23), CIN1 (41.93±9.10), CIN2 (42.37±9.71), CIN3 (40.77±8.09), CC (49.73±8.42) | RT-qPCR, Mir-Let-7 | miRNA-21, Mir-214, Mir-34a, Mir-200a | Mir-21 | Mir-214, Mir-200a, Mir -34a | Mir-214, Mir-200a, Mir -34a | |||||||
|
|
|||||||||||||||
| 7. Microrna-466 with tumour markers for cervical cancer screening | Zhou et al. [44], 2017, China, cohort study | 210 Normal, 280 cervical hyperplasia, 143 SCC, 102 AD, serum | Median: 49 | RT-qPCR, U6snRNA | Mir-466 | Mir-466 | |||||||||
|
|
|||||||||||||||
| 8. Expression profile of_microRNA-203_and Its__np63 target In cervical carcinogenesis: prospects for cervical cancer screening | Piersma et al. [34], 2016, Brazil, cross-sectional study | 21 Normal, 18 CIN I, 20 CIN II, 21 CIN III, 15 CC, fresh tissue | N/A | RT-qPCR, Mie-191, Mir-23a | Mir-203 | Mir-203 | Mir-203 | Mir-203 | Mir-203 | ||||||
|
|
|||||||||||||||
| 9. Novel microRNA signatures in HPV-mediated cervical carcinogenesis in Indian women | Sharma et al. [23], 2016, India, case-control study | 30 Normal, 20 CIN, 50 CC, frozen tissue | N/A | RT-qPCR, U6 snRNA, RNU43, RtU snRNA | Mir-200b, Mir-146a, Mir-3130-5p, Mir-95, Mir-136, Mir-34a, Let-7a, Mir-23b, Mir-29, Mir-19a, Mir-222, Mir-1, Mir-29a, Mir-576-3, Mir-21, Mir-449b, Mir-184, Mir-17, Mir-99a, Mir-376b, Mir-193-5p, Mir-423-5p, Mir-15a, Mir-517a, Mir-545, Mir-223, Mir-192 | Mir-15a, Mir-449b, Mir-517a, Mir-545, Mir-223, Mir-192 | Mir-34a, Mir-23b, Mir-99a, Mir-376b, Mir-193-5p, Mir-423-5p | Mir-29, Mir-19a, Mir-222, Mir-1, Mir-29a, Mir-222 and Mir-19a, Mir-576-3, Mir-21, Mir-449b, Mir-184, Mir-17 | Mir-200b, Mir-146a, Mir-3130-5p, Mir-95 and Mir-136, Mir-34a, Let-7a, Mir-23b | ||||||
|
|
|||||||||||||||
| 10. Up-regulation of mir-21 is associated with cervicitis and human papillomavirus infection in cervical tissues | Bumrungthai et al. [27], 2015, Thailand, cross-sectional study | 20 Normal, 12 cervicitis, 14 CIN I, 22 CIN II-III, and 43 SCC, fresh tissue and exfoliated cells | N/A | RT-qPCR, U6snRNA | Mir-21 | Mir-21 | Mir-21 | ||||||||
|
|
|||||||||||||||
| 11. Mir-34a and mir-125b expression in HPV infection and cervical cancer development | Ribeiro et al. [20], 2015, Portugal, cross-sectional study | 49 Normal, 28 LSIL, 29 HSIL, 8 ICC, exfoliated cells | Mean: 40±12.6 | RT-qPCR, Mir-23a | Mir-34a, Mir-125b | Mir-34a | Mir-125b | ||||||||
|
|
|||||||||||||||
| 12. Detection of high-grade neoplasia in air-dried cervical PAP smears by a microRNA-based classifier | Ivanov et al. [29], 2018, Russia, cross-sectional study | 40 Normal, 34 LSIL, 57 HSIL, 43 ICC, exfoliated cells | Mean: normal (31), LSIL (36), HSIL (44), ICC (53) | RT-qPCR, U6snRNA | Mir-106b, Mir-1246, Mir-126, Mir-196b, Mir-20a, Mir-21, Mir-375, Mir-145 | Mir-375, Mir-145 | Mir-106, Mir-1246, Mir-126, Mir-196b, Mir-20a, Mir-21 | Mir-375, Mir-145 | Mir-106b, Mir-1246, Mir-126, Mir-196b, Mir-20a, Mir-21 | ||||||
|
|
|||||||||||||||
| 13. Mir-21-5p, Mir-34a, and human telomerase RNA component as surrogate markers for cervical cancer progression | Zhu et al. [53], 2018, China, case-control study | 18 NILM, 45 ASCUS, 21 LSIL, 25 HSIL, 9 SCC, fresh tissue and exfoliated cells | Mean: 43.5±8.5 | RT-qPCR, U6snRNA | Mir-21-5p, Mir-34a | Mir-21-5p, | Mir-34a | Mir-21-5p | Mir-34a | Mir-21-5p | Mir-34a | Mir-21-5p | Mir-34a | Mir-21-5p | Mir-34a |
|
|
|||||||||||||||
| 14. Identification of miRNAs in cervical mucus as A novel diagnostic marker for cervical neoplasia | Kawai et al. [28], 2018, Japan, cross-sectional study | 56 Normal, 19 CIN I, 33 CIN II, 43 CIN III, 35 SCC, 19 AD, mucus and exfoliated cells | Median: normal (36), CIN I (39), CIN II (38), CIN III (36), SCC (54), AD (47) | Microarray analysis and validated by RT-qPCR, RNU48 | Mir-126-3p, Mir -20b-5p, Mir-451a, Mir-144-3p | Mir-126-3p, Mir-20b-5p, Mir-451a, Mir-144-3p | Mir-126-3p, Mir-20b-5p, Mir-451a, Mir-144-3p | Mir-144-3p | |||||||
|
|
|||||||||||||||
| 15. Triage of high-risk HPV-positive women in population-based screening by miRNA expression analysis in cervical scrapes; a feasibility study | Babion et al. [16], 2018, the Netherlands, case-control study | 74 Normal, 139 CIN II-III, 51 SCC, 20 AD, cervical scrapes and frozen tissue | Median: normal (41), CIN III (35), SCC (51) AD (45) | RT-qPCR, RNU24, Mir-423-3p | Mir-9-5p, Mir-149-5p, Mir-203a-3p, Mir-125b-5p, Mir-15b-5p, Mir-375 | Mir-9-5p, Mir-15b-5p | Mir-149-5p, Mir-203a-3p, Mir-375, Mir-125b-5p | Mir-9-5p, Mir-15b-5p | Mir-149-5p, Mir-203a-3p, Mir-125b-5p, Mir-375 | Mir-15b-5p | Mir-149-5p, Mir-375 | ||||
|
|
|||||||||||||||
| 16. Mir-1266 promotes cell proliferation, migration and invasion in cervical cancer by targeting DAB2IP | Wang et al. [43], 2018, China, cohort study | 100 Normal, 50 LSIL, 50 HSIL, 100 CC, serum and frozen tissue | N/A | RT-qPCR, U6snRNA | Mir-1266 | Mir-1266 | Mir-1266 | Mir-1266 | |||||||
|
|
|||||||||||||||
| 17. miRNA detection in cervical exfoliated cells for missed high-grade lesions in women with LSIL/ CIN1 diagnosis after colposcopy-guided biopsy | Ye et al. [40], 2019, China, cross-sectional study | 150 LSIL, 27 HSIL, exfoliated cells and tissue | Mean: LSIL (39.4±6.6), HSIL (39.8±7.1) | RT-qPCR, U6snRNA | miRNA-195, miRNA-29a, miRNA-16- 2, miRNA-20a | miR-NA-16-2, miRNA-20a | miR-NA-195, miRNA-29a | ||||||||
|
|
|||||||||||||||
| 18. Increased expression of Mir-15b is associated with clinicopathological features and poor prognosis in cervical carcinoma | Wen et al. [46], 2017, China, cohort Study | 150 Normal, 124 CIN I, 148 CIN II-III, 185 CC, frozen tissue | Mean: normal (49.23±7.68), CIN I -III (48.60±4.2), CC (49.07±8.52) | RT-qPCR, U6snRNA | Mir-15b | Mir-15b | Mir-15b | Mir-15b | Mir-15b | Mir-15b | |||||
|
|
|||||||||||||||
| 19. Reduced Mir-34a expression in normal cervical tissues and cervical lesions with high-risk human papillomavirus infection | Li et al. [55], 2010, China, case-control study | 64 Normal, 44 CIN I -III, 32 CC, fresh tissue | N/A | RT-PCR and gel electrophoresis, β-actin | Pri-Mir-34a | Pri-Mir-34a | Pri-Mir-34a | Pri-Mir-34a | |||||||
|
|
|||||||||||||||
| 20. Mir-196a targets netrin 4 and regulates cell proliferation and migration of cervical cancer cells | Zhang et al. [41], 2013, China, cross-sectional study | 24 Normal, 13 CIN I, 18 CIN II-III, 15 SCC, frozen tissue | N/A | RT-qPCR, U6snRNA | Mir-196a | Mir-196a | Mir-196a | Mir-196a | |||||||
|
|
|||||||||||||||
| 21. Mir-3156-3p is downregulated in HPV-positive cervical cancer and performs as a tumour-suppressive miRNA | Xie et al. [33], 2017, China, case-control study | 40 Normal, 50 CC, frozen tissue | N/A | RT-qPCR, U6snRNA | Mir-3156-3p | Mir-3156-3p | |||||||||
|
|
|||||||||||||||
| 22. The role of mir-409-3p in regulation of HPV16/18-E6 mRNA in human cervical high-grade squamous intraepithelial lesions | Sommerova et al. [32], 2019, Czech republic, case-control study | 54 Normal, 90 HSIL, FFPE tissue | Median: 36 | RT-qPCR, U6snRNA | Mir-10a-5p, Mir-132-3p, Mir-141-5p, Mir-10b-5p, Mir-34c-5p, Mir-409-3p, Mir-411-5p | Mir-10a-5p, Mir-132-3p, Mir-141-5p | Mir-10b-5p, Mir-34c-5p, Mir-409-3p, Mir-411-5p | ||||||||
|
|
|||||||||||||||
| 23. MicroRNA expression variability in human cervical tissues | Pereira et al. [21], 2010, Portugal, cross-sectional study | 19 Normal, 9 LSIL, 7 HSIL, 4 SCC, frozen tissue | Range: 21-51 | Microarray analysis and validated By RT-qPCR, RNU6B | Mir-26a, Mir-143, Mir-145, Mir-99a, Mir-203, Mir-513, Mir-29a, Mir-199a, Mir-106a, Mir-205, Mir-197, Mir-16, Mir-27a, Mir-142-5p, Mir-148a, Mir-302b, Mir-10a, Mir-196a, Mir-132, Mir-522, Mir-512-3p | Mir-148a, Mir-302b, Mir-10a, Mir-196a, Mir-132 | Mir-26a, Mir-143, Mir-145, Mir-99a, Mir-203, Mir-513, Mir-29a, Mir-199a | Mir-522 and Mir-512-3p, Mir-148a, Mir-302b, Mir-10a, Mir-196a, Mir-132 | Mir-26a, Mir-143, Mir-145, Mir-99a, Mir-203, Mir-513, Mir-29a, Mir-199a, Mir-106a, Mir-205, Mir-197, Mir-16, Mir-27a, Mir-142-5p | Mir-106a, Mir-205, Mir-197, Mir-16, Mir-27a and Mir-142-5p, Mir-148a, Mir-302b, Mir-10a, Mir-196a, Mir-132 | Mir-26a, Mir-143, Mir-145, Mir-99a, Mir-203, Mir-513, Mir-29a, Mir-199a, Mir-522, Mir-512-3p | ||||
|
|
|||||||||||||||
| 24. Expression of Mir200a, Mir93, metastasis-related gene RECK and MMP2/ MMP9 in human cervical carcinoma-relationship with prognosis | Wang et al. [50], 2013, China, cohort study | 100 Normal, 99 SCC, 17 AD frozen tissue | Mean: 49.3±2.39 | RT-qPCR, U6snRNA | Mir93, Mir200a | Mir93, Mir200a | |||||||||
|
|
|||||||||||||||
| 25. Alterations in microRNAs Mir-21 and let-7a correlate with aberrant STAT3 signaling and downstream effects during cervical carcinogenesis | Shishodia et al. [24], 2015, India, cross-sectional study | 23 Normal, 10 LSIL, 13 HSIL, 56 CC, fresh tissue | N/A | RT-qPCR, U6snRNA | Mir-21, Let-7a | Mir-21 | Let-7a | Mir-21, Let-7a | Mir-21 | Mir-21 | Let-7a | ||||
|
|
|||||||||||||||
| 26. Interferon-β induced microRNA--129-5p down-regulates HPV-18 E6 and E7 viral gene expression by targeting SP1 in cervical cancer cells | Wang et al. [43], 2019, China, cross-sectional study | 4 Normal, 6 CIN I-II, 13 CIN III, 14 CC, frozen tissue | Range: 28-77 | Microarray analysis and validated by RT-qPCR, RNU6B | Mir-129-5p | Mir-129-5p | Mir-129-5p | Mir-129-5p | Mir-129-5p | Mir-129-5p | |||||
|
|
|||||||||||||||
| 27. Dysregulated microRNAs involved in the progression of cervical neoplasm | Zeng et al. [51], 2015, China, cross-sectional study | 16 Normal, 18 LSIL, 38 HSIL, 43 SCC, FPPE tissue | Mean: Normal (46.65±4.46), LSIL (42.80±10.03), HSIL (45.65±10.22), SCC (50.08±8.47) | Microarray analysis and validated by RT-qPCR, U6snRNA | Mir-21, Mir-218, Mir-376a, Mir-31, Mir-9, Mir-195, Mir-497, Mir-199b-5p | Mir-21, Mir-31, Mir-9 | Mir-376a, Mir-218, Mir-195, Mir-497, Mir-199b-5p | Mir-218, Mir-195, Mir-497, Mir-199b-5p | |||||||
|
|
|||||||||||||||
| 28. Altered microRNA expression associated with chromosomal changes contributes to cervical carcinogenesis | Wilting et al. [19], 2013, the Netherlands, cross-sectional study | 10 Normal, 18 CIN II-III, 10 SCC, 9 AD, frozen tissue | Range: 30-72 | Microarray analysis and validated by RT-qPCR, RNU43 | Mir-9, Mir-15b, Mir-28-5p, Mir-21, Mir-100, Mir-125b, Mir-375, Mir-203 | Mir-9, Mir-15b, Mir-28-5p, Mir-21 | Mir-100, Mir-125b, Mir-375, Mir-203 | Mir-28-5p, Mir-21 | Mir-203 | Mir-9, Mir-15b, Mir-28-5p, Mir-21 | Mir-100, Mir-125b, Mir-375, Mir-203 | ||||
|
|
|||||||||||||||
| 29. Let-7c is a candidate biomarker for cervical intraepithelial lesions: a pilot study | Malta et al. [22], 2015, Portugal, case-control study | 38 Normal, 14 LSIL, 21 HSIL, exfoliated cells | Mean: 40±13.0 | RT-qPCR, Mir-23a | Let-7c | Let-7c | |||||||||
|
|
|||||||||||||||
| 30. Folate inhibits Mir-27a-3p expression during cervical carcinoma progression and oncogenic activity in human cervical cancer cells | Wang et al. [49], 2020, China, case-control study | 30 Normal, 30 HSIL, 40 SCC, frozen tissue | Mean: sufficient folate group (47.40±2.41), insufficient folate group (48.40±1.82) | Microarray analysis and validated by RT-qPCR, U6snRNA | Mir-27a-3p | Mir-27a-3p | Mir-27a-3p | Mir-27a-3p | |||||||
|
|
|||||||||||||||
| 31. MicroRNAs are biomarkers of oncogenic human papillomavirus infections | Wang et al. [48], 2014, China, case-control study | 38 Normal, 13 CIN I-II, 39 CIN III, 68 CC, FFPE tissue | N/A | Microarray analysis and validated by RT-qPCR, U6snRNA | Mir-378, Mir-27a | Mir-378, Mir-27a | Mir-378, Mir-27a | Mir-27a | |||||||
|
|
|||||||||||||||
| 32. Evaluation of microRNA-205 expression as a potential triage marker for patients with low-grade squamous intraepithelial lesions | Xie et al. [33], 2017, Sweden, cross-sectional study | 16 Normal, 29 CIN I, 44 CIN II, 47 CIN III, 4 CC, exfoliated cells | Median: 32.5 | RT-qPCR, RNU6B | No significance | ||||||||||
|
|
|||||||||||||||
| 33. Complementarity between miRNA expression analysis and DNA methylation analysis in hr-HPV positive cervical scrapes for the detection of cervical disease | Babion et al. [17], 2019, The Netherlands, case-control study | 52 Normal, 25 CIN I, 48 CIN II, 55 CIN III, 24 SCC, 5 AD, exfoliated cells | Median: normal (39.5), CIN I (35.0), CIN II (34.5), CIN3 (35.0), SCC (48.5), AD (50.0) | RT-qPCR, RNU24, Hsa-Mir-423-3p | Mir-15b, Mir-125b, Mir-149, Mir-203a, Mir-375, Let-7b, Mir-93, Mir-222 | Mir-15b, Mir-125b, Mir-149, Mir-203a, Mir-375, Let-7b, Mir-93, Mir-222 | |||||||||
|
|
|||||||||||||||
| 34. Methylation and expression of miRNAs in precancerous lesions and cervical cancer with HPV16 infection | Jiménez-Wences et al. [25], 2016, Mexico, cross-sectional study | 17 Normal, 16 LSIL, 11 CC, exfoliated cells and fresh tissue | Mean: normal HPV-(32.2), normal HPV+ (29.9), LSIL (29.9), CC (52.8) | RT-qPCR, Mir-92a | Mir-124, Mir-218, Mir-193b | Mir-193b | Mir-124, Mir-218 | ||||||||
|
|
|||||||||||||||
| 35. Mir-23b as a potential tumor suppressor and Its regulation by DNA methylation in cervical cancer | Campos-Viguri et al. [26], 2015, Mexico, cross-sectional study | 18 Normal, 19 LSIL, 7 HSIL, 28 CC, exfoliated cells and fresh tissue | N/A | RT-qPCR, Mir-92a | Mir-23b | Mir-23b | Mir-23b | ||||||||
|
|
|||||||||||||||
| 36. Methylation-mediated silencing and tumour suppressive function of hsa-mir-124 in cervical cancer | Wilting et al. [18], 2010, the Nether-lands, cross-sectional study | 23 Normal, 36 CIN I, 48 CIN II-III, 38 SCC and 20 AD, exfoliated cells, frozen tissue and FFPE tissue | Mean: 40.3 | RT-qPCR, RNU43 | Mir-124 | Mir-124 | Mir-124 | ||||||||
|
|
|||||||||||||||
| 37. A correlational study on Mir-34s and cervical lesions | Liu et al. [39], 2018, China, case-control study | 30 Normal, 30 CIN1, 30 CIN2/3, 30 CC, frozen tissue | Mean: 45.77±10.73 | RT-qPCR, U6snRNA | miRNA-34 | miRNA-34 | |||||||||
|
|
|||||||||||||||
| 38. Clinical value of combined detection of Mir-1202 and Mir-195 in early diagnosis of cervical cancer | Yang et al. [45] 2019, China, case-control study | Mean: normal (41.1±2.8), CC (40.2±3.7) | RT-qPCR, U6snRNA | Mir-1202, Mir-195 | Mir-1202, Mir-195 | ||||||||||
CC, cervical cancer; CIN, cervical intraepithelial neoplasia; LSIL, low-grade squamous intraepithelial lesion; HSIL, high-grade squamous intraepithelial lesion; NILM, negative for intraepithelial lesion or malignancy; FFPE, formalin-fixed paraffin embedded; RT-qPCR, quantitative reverse transcription polymerase chain reaction; PDK, pyruvate dehydrogenase kinase; N/ A, not available; ASCUS, atypical squamous cells of undetermined significance; SCC, squamous cell carcinoma; HPV, human papillomavirus; AD, adenocarcinoma; ICC, intrahepatic cholangiocarcinoma; PAP, poly A polymerase; DAB2IP, disabled homolog 2-interacting protein; RECK, reversion-inducing-cysteine-rich protein; MMP, matrix metalloproteinases.
References
- TOOLS
-
METRICS

- Related articles in Obstet Gynecol Sci
-
Social determinants of mental health of women living in slum: a systematic review2021 March;64(2)